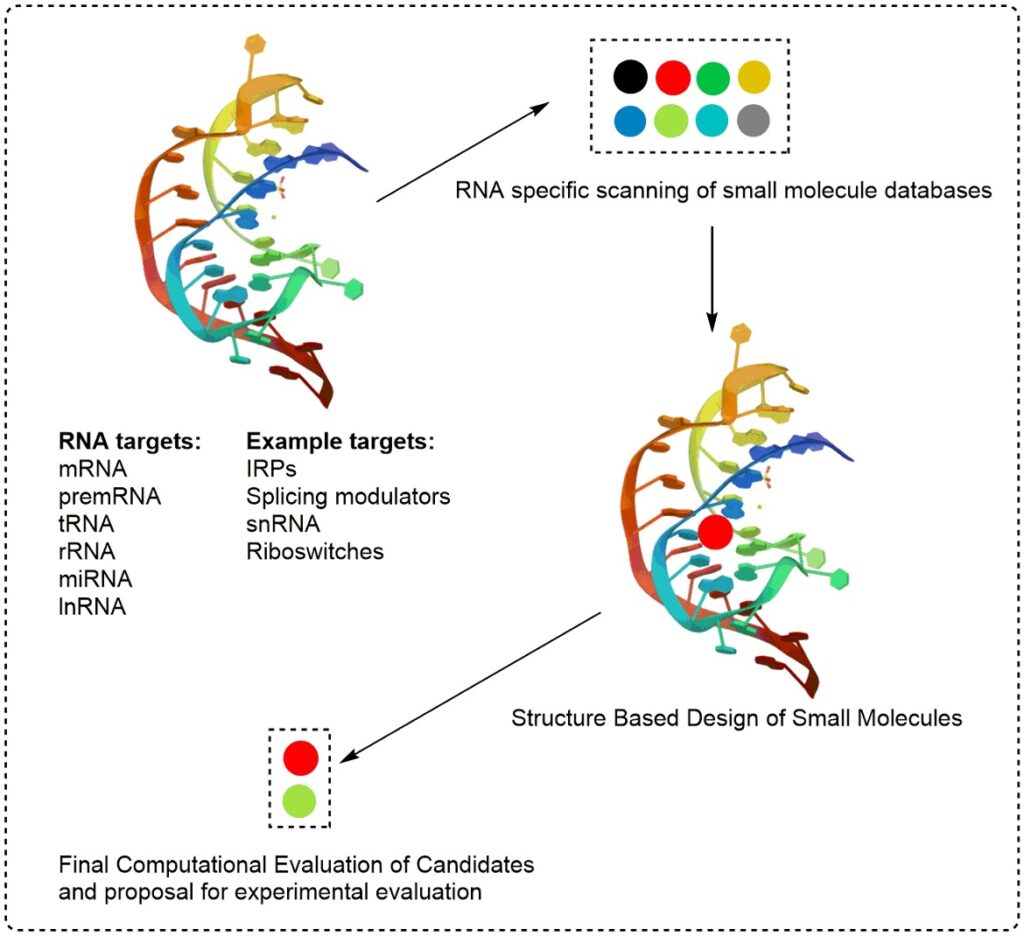
- Small Molecule Drug Design
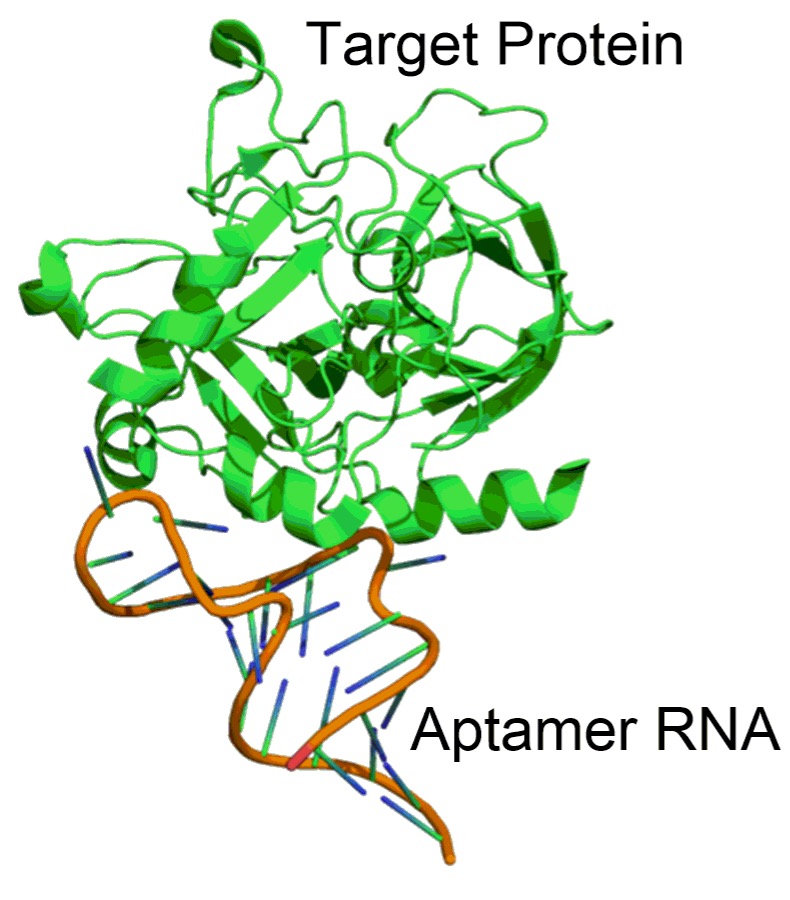
- Therapeutic RNA Aptamers
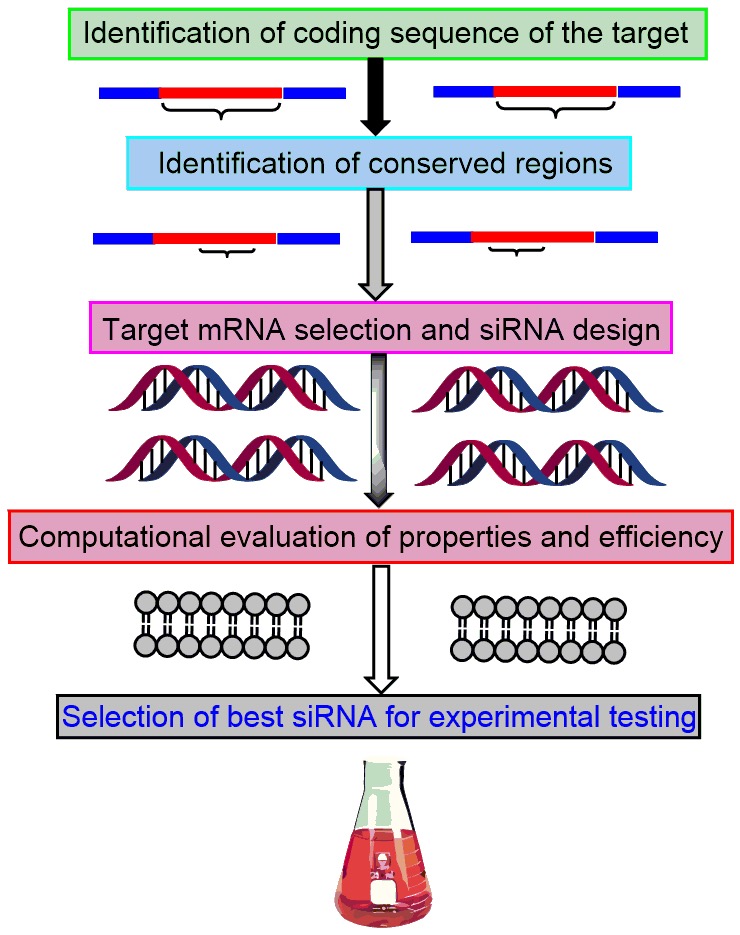
- Computational siRNA Design
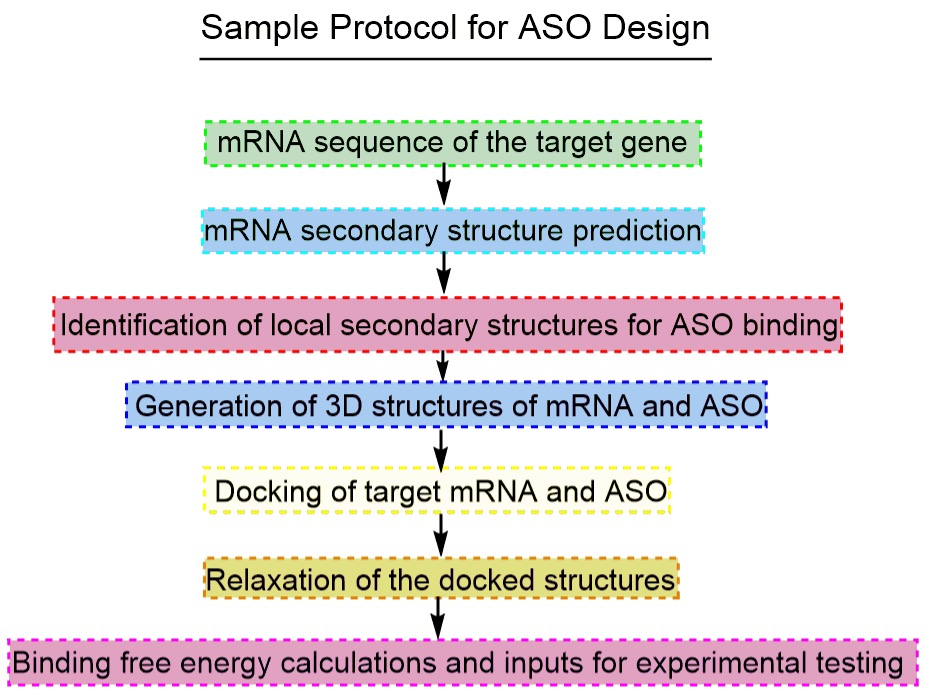
- Computational Design of Antisense Oligonucleotides
More commonly employed for protein targets, structure-based design and molecular docking have enabled the discovery and the optimization of lead compounds for RNA targets. Small molecule libraries can be docked to well defined RNA structural ensembles to enable in silico screening for selective RNA binding.
Aptamers are RNA oligonucleotides that bind to a specific target with high affinity, and specificity and can thus be used to target therapeutically important proteins. Major efforts have recently been undertaken to create aptamers for a wide range of diseases using the experimentally laborious SELEX (systematic evolution of ligands by exponential enrichment). However, our robust computational aptamer design framework, involving construction of the aptamer structures, predicting their binding conformations, and optimizing their binding through mutations, is very useful in accelerating RNA aptamer-based therapeutics.
siRNAs represent the next-generation molecular therapeutics capable of treating a variety of diseases through gene silencing. siRNAs can play a central role in inhibiting the expression of disease-causing genes by hybridization and subsequent inactivation of the complementary target mRNAs through the RNA interference (RNAi) pathway. We employ computational tools to design experimentally testable specific siRNA sequences for different targets.
ASOs are an important class of drugs that function through interfering with mRNA processing through RNase H–mediated degradation, translational arrest, or splicing modulation. Several factors determine the binding efficiency of the ASO to the target sequences. Our robust computational protocol help design efficient ASOs for experimental testing.