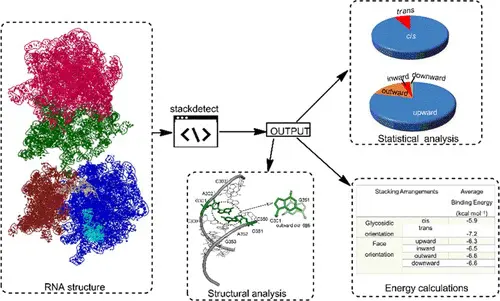
Our paper on statistical analysis and Quantum Chemical Calculation on RNA base stacks is now available on JCIM website. This fundamental contribution to understanding of RNA structures will hopefully help design better RNA folding algorithms and in the field of RNA-based drug design.
Nucleobase π–π stacking is one of the crucial organizing interactions within three-dimensional (3D) RNA architectures. Characterizing the structural variability of these contacts in RNA crystal structures will help delineate their subtleties and their role in determining function. This analysis of different stacking geometries found in RNA X-ray crystal structures is the largest such survey to date; coupled with quantum-mechanical calculations on typical representatives of each possible stacking arrangement, we determined the distribution of stacking interaction energies. A total of 1,735,481 stacking contacts, spanning 359 of the 384 theoretically possible distinct stacking geometries, were identified. Our analysis reveals preferential occurrences of specific consecutive stacking arrangements in certain regions of RNA architectures. Quantum chemical calculations suggest that 88 of the 359 contacts possess intrinsically stable stacking geometries, whereas the remaining stacks require the RNA backbone or surrounding macromolecular environment to force their formation and maintain their stability. Our systematic analysis of π–π stacks in RNA highlights trends in the occurrence and localization of these noncovalent interactions and may help better understand the structural intricacies of functional RNA-based molecular architectures.