Our computational work on the interaction of Guanidinium salts with proteins is now available on PCCP’s website. This work will help better understand protein ligand binding and protein denaturation processes.
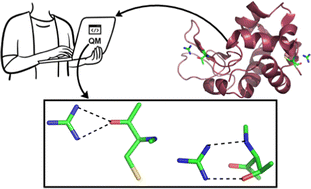
In the present work, 86 available high resolution X-ray structures of proteins that contain one or more guanidinium ions (Gdm+) are analyzed for the distribution and nature of noncovalent interactions between Gdm+ and amino-acid residues. A total of 1044 hydrogen-bonding interactions were identified, of which 1039 are N–H⋯O, and five are N–H⋯N. Acidic amino acids are more likely to interact with Gdm+ (46% of interactions, 26% Asp and 20% Glu), followed by Pro (19% of interactions). DFT calculations on the identified Gdm+–amino acid hydrogen-bonded pairs reveal that although Gdm+ interacts primarily with the backbone amides of nonpolar amino acids, Gdm+ does interact with the sidechains of polar and acidic amino acids. We classified the optimized Gdm+–amino acid pairs into parallel [p], bifurcated [b], single hydrogen bonded [s] and triple hydrogen bonded [t] types. The [p] and [t] type pairs possess higher average interaction strength that is stronger than that of [b] and [s] type pairs. Negatively charged aspartate and glutamate residues interact with Gdm+ ion exceptionally tightly (−76 kcal mol−1) in [p] type complexes. This work provides statistical and energetics insights to better describe the observed destabilization or denaturation process of proteins by guanidinium salts.